This is featured post 1 title
Replace these every slider sentences with your featured post descriptions.Go to Blogger edit html and find these sentences.Now replace these with your own descriptions.This theme is Bloggerized by Lasantha - Premiumbloggertemplates.com.

This is featured post 2 title
Replace these every slider sentences with your featured post descriptions.Go to Blogger edit html and find these sentences.Now replace these with your own descriptions.This theme is Bloggerized by Lasantha - Premiumbloggertemplates.com.

This is featured post 3 title
Replace these every slider sentences with your featured post descriptions.Go to Blogger edit html and find these sentences.Now replace these with your own descriptions.This theme is Bloggerized by Lasantha - Premiumbloggertemplates.com.

Saturday, November 27, 2010
Rasmol:gcufbioinfo
 10:16 AM
10:16 AM
 GCbioinfo
GCbioinfo
Rasmol:gcufbioinfo
 10:01 AM
10:01 AM
 GCbioinfo
GCbioinfo
click Display>and select different options to view your molecule.
Rasmol:gcufbioinfo
 9:46 AM
9:46 AM
 GCbioinfo
GCbioinfo
And save this file to any directory with file format ".pdb" so now you can visualize already saved molecule.
Rasmol:gcufbioinfo
 9:34 AM
9:34 AM
 GCbioinfo
GCbioinfo
enter protein name for which you want to view the structure. As I entered DNA Polymerase so the result is http://www.pdb.org/pdb/explore/explore.do?structureId=3OGU
the out put is as:
At the right of this page you can there is the option of Download files so click this. After clicking this select "PDB files(gz)".then download and install winzip to your system and download this file automatically to raswin and view the structure in rasmol.
What kinds of molecules can RasMol display?:Gcufbioinfo
 9:25 AM
9:25 AM
 GCbioinfo
GCbioinfo
In order to display a molecule, RasMol needs a data file called an atomic coordinate file. This data file specifies the position of every atom in the molecule, as cartesian coordinates X, Y, and Z.
Three-dimensional structures can be predicted for many small molecules, but must be determined empirically for macromolecules. The most common method for determining structure is X-ray diffraction analysis of a crystal. Nuclear magnetic resonance (NMR) can also be used. Some structures are available only as theoretical models, often based on related molecules for which empirical structures have been determined.
Rasmol Tool:
 8:19 AM
8:19 AM
 GCbioinfo
GCbioinfo
here. Install Rasmol to your system and get started.
Thursday, November 25, 2010
HTML Calculator:gcufbioinfo
 8:25 PM
8:25 PM
 GCbioinfo
GCbioinfo
function sqrt(form)
{
form.displayvalue.value=Math.sqrt(form.displayvalue.value);
}
function sq(form)
{
form.displayvalue.value=eval(form.displayvalue.value)*eval(form.displayvalue.value);
}
function ln(form)
{
form.displayvalue.value=Math.exp(form.displayvalue.value);
}
function equal(form)
{
form.displayvalue.value=eval(form.displayvalue.value);
}
function delChar(input)
{
input.value=input.value.substring(0,input.value-length-1);
}
function addChar(input,character)
{
if(input.value==null||input.value=="0")
input.value=character
else
input.value+=character
}
Insha Allah you will see your functioning very soon regards Admin Quratt ul ain Siddique.
Monday, November 22, 2010
HTML Calculator:gcufbioinfo
 8:53 AM
8:53 AM
 GCbioinfo
GCbioinfo
<head>
<script type="text/javascript">
function cos(form)
{
form.displayvalue.value=Math.cos(form.displayvalue.value);
}
function sin(form)
{
form.displayvalue.value=Math.sin(form.displayvalue.value);
}
function tan(form)
{
form.displayvalue.value=Math.tan(form.displayvalue.value);
}
</script>
</head>
I will tell you other codes as soon as possible and you will see Insha Allah your calculator functional
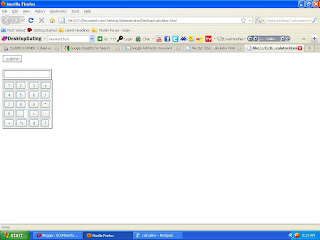
HTML Calculator:gcufbioinfo
 8:33 AM
8:33 AM
 GCbioinfo
GCbioinfo
<html>
<title>Calculator</title>
<head></head>
<body>
<table border="2">
<tr>
<td><input name="displayvalue" size="39" value="0"></td>
<tr>
<td>
<table border="2">
<tr>
<td>
<input type="button" value=" char ">
</td>
<td>
<input type="button" value=" delete ">
</td>
<td>
<input type="button" value=" = ">
</td>
</tr>
</table>
</td>
<tr>
<td>
<table border="2">
<tr>
<td>
<input type="button" value=" exp ">
</td>
<td>
<input type="button" value=" 7 ">
</td>
<td>
<input type="button" value=" 8 ">
</td>
<td>
<input type="button" value=" 9 ">
</td>
<td>
<input type="button" value=" / ">
</td>
</tr>
<tr>
<td>
<input type="button" value=" ln ">
</td>
<td>
<input type="button" value=" 4 ">
</td>
<td>
<input type="button" value=" 5 ">
</td>
<td>
<input type="button" value=" 6 ">
</td>
<td>
<input type="button" value=" * ">
</td>
</tr>
<tr>
<td>
<input type="button" value=" sqrt ">
</td>
<td>
<input type="button" value=" 1 ">
</td>
<td>
<input type="button" value=" 2 ">
</td>
<td>
<input type="button" value=" 3 ">
</td>
<td>
<input type="button" value=" - ">
</td>
</tr>
<tr>
<td>
<input type="button" value=" sq ">
</td>
<td>
<input type="button" value=" 0 ">
</td>
<td>
<input type="button" value=" . ">
</td>
<td>
<input type="button" value=" % ">
</td>
<td>
<input type="button" value=" + ">
</td>
</tr>
<tr>
<td>
<input type="button" value=" ( ">
</td>
<td>
<input type="button" value=" cos ">
</td>
<td>
<input type="button" value=" sin ">
</td>
<td>
<input type="button" value=" tan ">
</td>
<td>
<input type="button" value=" ) ">
</td>
</tr>
</table>
</td>
</tr>
</tr>
</tr>
</table>
</body>
</html>
out put is as:
Now I tell you how to make it functional
Quratt ul ain Siddique
Sunday, November 21, 2010
imageJ:gcufbioinfo
 5:57 AM
5:57 AM
 GCbioinfo
GCbioinfo
Medical Image Processing tool:
 5:46 AM
5:46 AM
 GCbioinfo
GCbioinfo
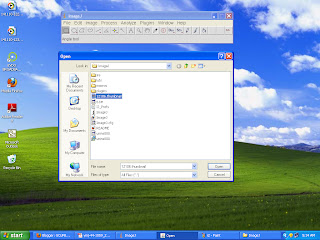
First save an image especially a medical image to your system and copy this image and go to C derive then program files and then imagej folder and paste this image into it. then open imageJ from start menu and click file >open as shown in gcufbioinfo and select the picture to view.
you can view your image using this tool .
imageJ:gcufbioinfo
 5:10 AM
5:10 AM
 GCbioinfo
GCbioinfo
Download ImageJ:
 4:50 AM
4:50 AM
 GCbioinfo
GCbioinfo
the page will open
click download
http://rsbweb.nih.gov/ij/download.html
then get latest version of imageJ for your OS . If you are new to use ImageJ then Download ImageJ 1.43 bundled with 32-bit Java 1.6.0_10 (25MB) other wise download without java for windows OS.
Medical Image Processing tool:
 4:37 AM
4:37 AM
 GCbioinfo
GCbioinfo
ImageJ is a public domain, Java-based image processing program developed at the National Institutes of Health. ImageJ was designed with an open architecture that provides extensibility via Java plugins and recordable macros. Custom acquisition, analysis and processing plugins can be developed using ImageJ's built-in editor and a Java compiler. User-written plugins make it possible to solve many image processing and analysis problems, from three-dimensional live-cell imaging, to radiological image processing,multiple imaging system data comparisonsto automated hematology systems.ImageJ's plugin architecture and built in development environment has made it a popular platform for teaching image processing.
Tuesday, November 2, 2010
primary structure analysis:
 8:11 AM
8:11 AM
 GCbioinfo
GCbioinfo
Radar:De novo repeat detection in protein sequences
I entered the protein albumin in uniprot and get the following result It gave the number of repeats present in the sequence.
http://www.ebi.ac.uk/Tools/es/cgi-bin/jobresults.cgi/radar/radar-20101102-1505421645.html
Monday, November 1, 2010
primary structure analysis
 7:49 AM
7:49 AM
 GCbioinfo
GCbioinfo
Radar :De novo repeat detection in protein sequences go to http://www.ebi.ac.uk/Tools/Radar/index.html
primary structure analysis:
 7:39 AM
7:39 AM
 GCbioinfo
GCbioinfo
HeliQuest A web server to screen sequences with specific alpha-helical properties HeliQuest calculates from an α-helix sequence its physicochemical properties and amino acid composition and uses the results to screen any databank in order to identify protein segments possessing similar features.
The server is divided into 2 interconnected modules: the sequence analysis module and the screening module.
In addition, the mutation module, (available from the sequence analysis module), allows user to mutate helices manually or automatically by genetic algorithm to create analogues with specific properties.-http://heliquest.ipmc.cnrs.fr/
After entering the helix sequence of the protein this protein sequence is get by entering the name of the protein in uniprot then paste it by clicking the analysis button and get the following
http://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParams.py
 7:19 AM
7:19 AM
 GCbioinfo
GCbioinfo
ScanSite pI/Mw:Compute the theoretical pI and Mw, and multiple phosphorylation states. Enter a protein name and sequence below. The molecular weight and isoelectric point of this sequence and multiple phosphorylation states will be displayed. http://scansite.mit.edu/calc_mw_pi.html
out put will be
primary structure analysis 3:
 7:06 AM
7:06 AM
 GCbioinfo
GCbioinfo
Compute pI/Mw: Compute the theoretical isoelectric point (pI) and molecular weight (Mw) from a UniProt Knowledgebase entry or for a user sequence
now go to this tool and get the required result Please enter one or more UniProtKB/Swiss-Prot protein identifiers (ID) (e.g. ALBU_HUMAN) or UniProt Knowledgebase accession numbers (AC) (e.g. P04406), separated by spaces, tabs or newlines. Alternatively, enter a protein sequence in single letter code. The theoretical pI and Mw (molecular weight) will then be computed. previosly I entered the amino acid sequence and now I am going to enter the uniprot AC#.http://www.expasy.org/cgi-bin/pi_tool
it will give you this format of result so click on the following things according to the instructions mentioned Please select one of the following features by clicking on a pair of endpoints, and the computation will be carried out for the corresponding sequence fragment.
Primary structure analysis 2:
 6:51 AM
6:51 AM
 GCbioinfo
GCbioinfo
ProtParam :Physico-chemical parameters of a protein sequence (amino-acid and atomic compositions, isoelectric point, extinction coefficient, etc.)
First go to this page http://www.expasy.org/tools/ select Primary structure analysis and select the first tool as I mentioned. click the tool and I entered the amino acid sequence of protein transgelin-2 in http://www.expasy.org/tools/protparam.html then get the out put page
http://www.expasy.org/cgi-bin/protparam
the out put is
Primary structure analysis:
 6:36 AM
6:36 AM
 GCbioinfo
GCbioinfo
I will breifly tell you how to use these tools.
HTML Calculator:
 6:22 AM
6:22 AM
 GCbioinfo
GCbioinfo
<input type="button" value="submit">
<html>
<body>
<input type="button" value="submit">
<br><br>
<table border="2">
<tr><td><input type="text"></td></tr>
<tr><table border="2"><tr><td><input type="button" value="1"></td><td><input type="button" value="2"></td><td><input type="button" value="3"></td><td><input type="button" value="c"></td></tr><tr><td><input type="button" value="4"></td><td><input type="button" value="5"></td><td><input type="button" value="6"></td><td><input type="button" value="/"></td></tr><tr><td><input type="button" value="7"></td><td><input type="button" value="8"></td><td><input type="button" value="9"></td><td><input type="button" value="*"></td></tr><tr><td><input type="button" value="0"></td><td><input type="button" value="."></td><td><input type="button" value="="></td><td><input type="button" value="-"></td></tr><tr><td><input type="button" value="+"></td><td><input type="button" value="%"></td><td><input type="button" value="d"></td><td><input type="button" value="f"></td></tr></table></tr>
</table>
</body>
</html>